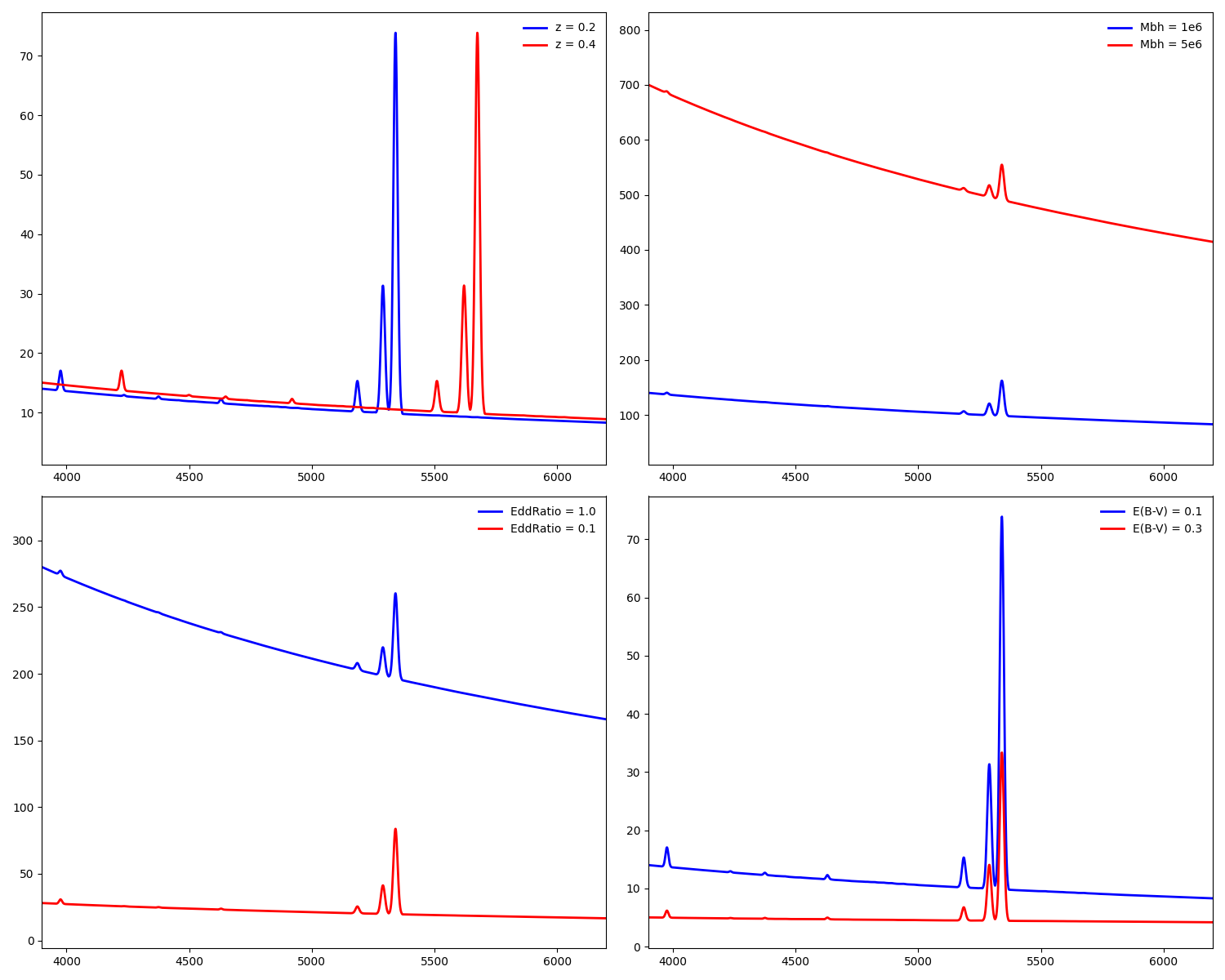
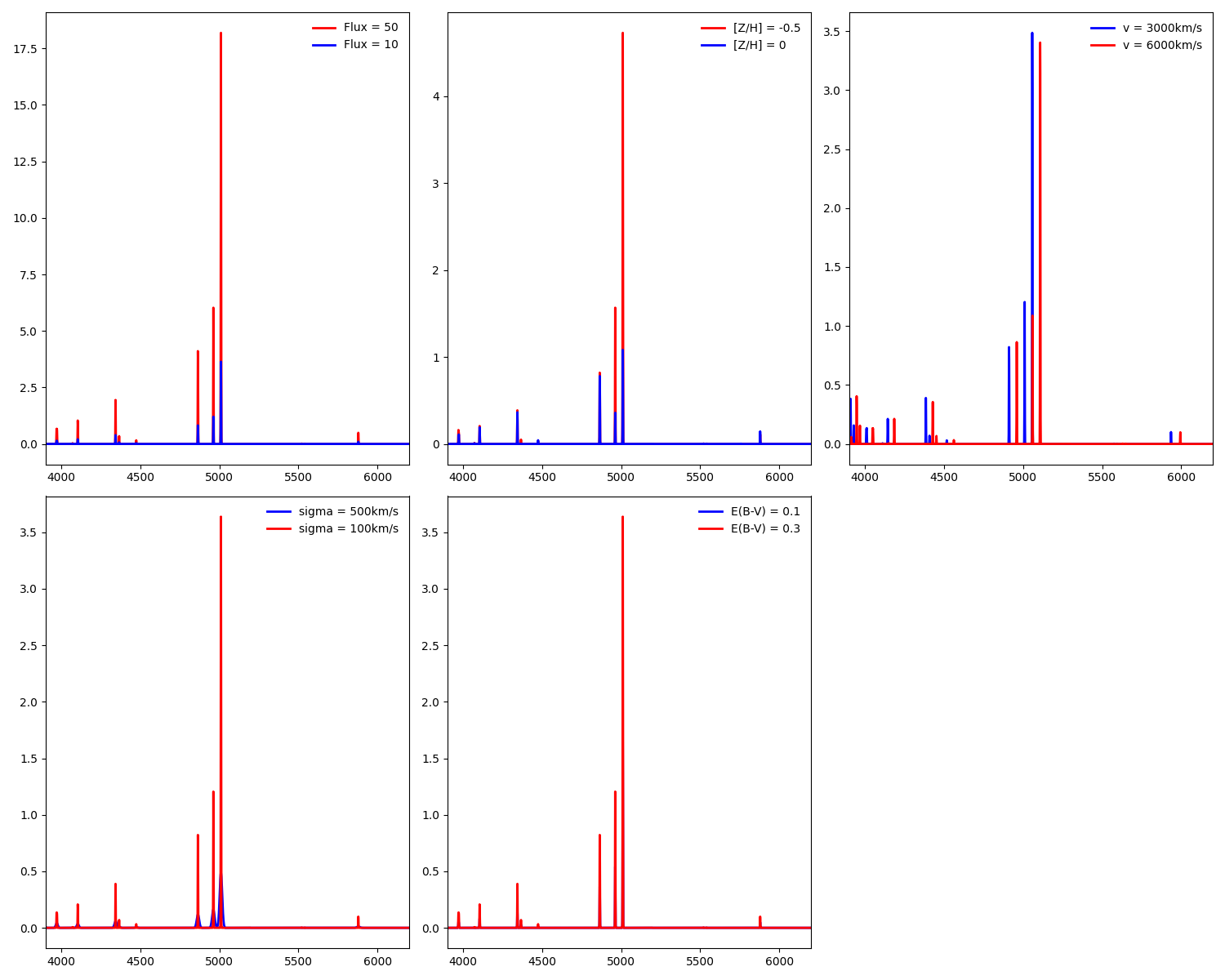
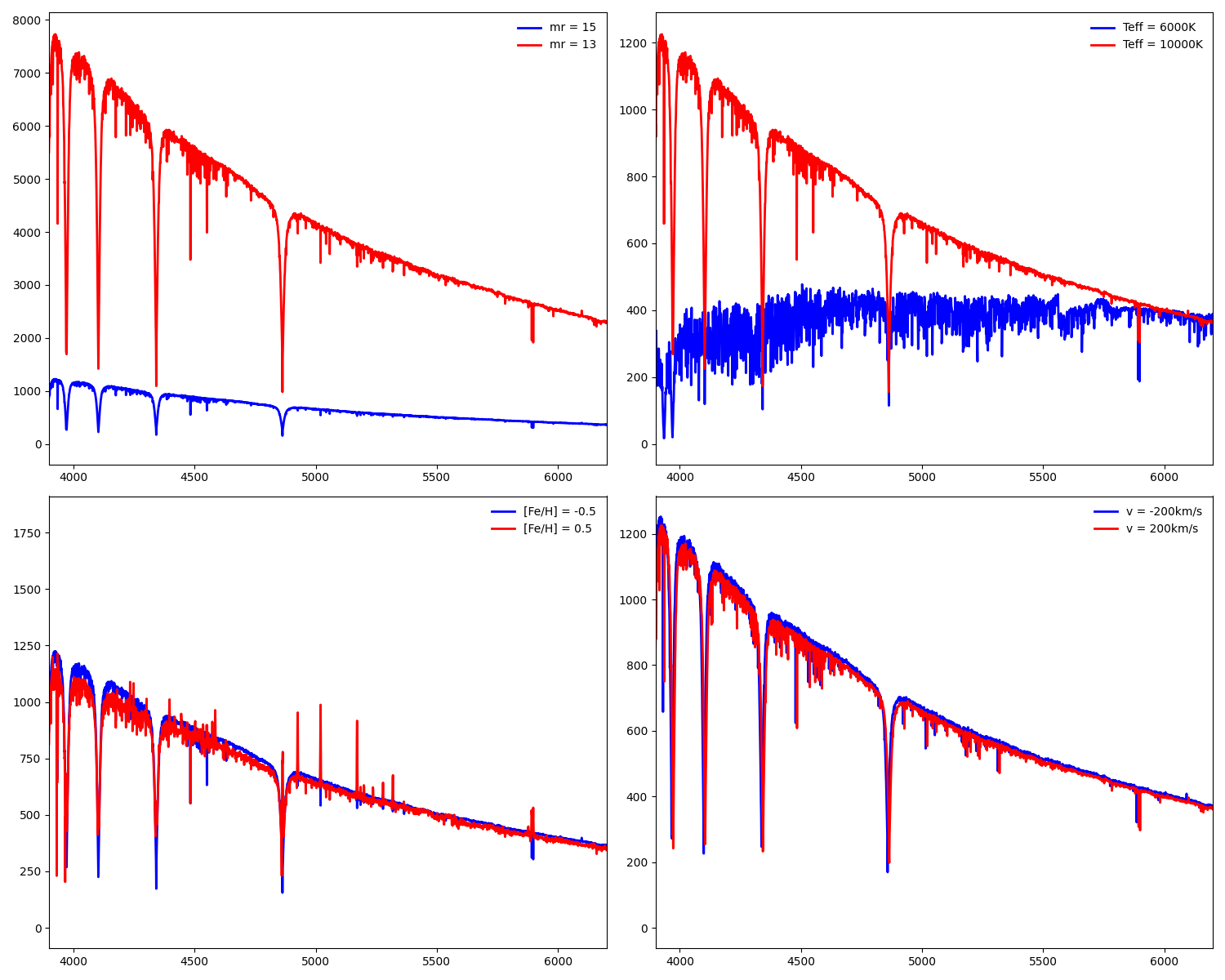
Deleted gehong/__pycache__/__init__.cpython-38.pyc,...
Deleted gehong/__pycache__/__init__.cpython-38.pyc, gehong/__pycache__/config.cpython-38.pyc, gehong/__pycache__/cube3d.cpython-38.pyc, gehong/__pycache__/map2d.cpython-38.pyc, gehong/__pycache__/spec1d.cpython-38.pyc, gehong/data/EMILES/model/HST_ch_iPp0.00.MAG, gehong/data/EMILES/model/HST_mag.dat, gehong/data/EMILES/model/out_mass_CH_PADOVA00, gehong/data/EMILES/.DS_Store, gehong/data/EMILES/Ech1.30Zm0.40T00.0631_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.40T00.0708_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.40T00.0794_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.40T00.0891_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.40T00.1000_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.40T00.1122_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.40T00.1259_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.40T00.1413_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.40T00.1585_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.40T00.1778_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.40T00.1995_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.40T00.2239_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.40T00.2512_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.40T00.2818_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.40T00.3162_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.40T00.3548_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.40T00.3981_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.40T00.4467_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.40T00.5012_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.40T00.5623_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.40T00.6310_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.40T00.7079_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.40T00.7943_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.40T00.8913_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.40T01.0000_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.40T01.1220_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.40T01.2589_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.40T01.4125_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.40T01.5849_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.40T01.7783_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.40T01.9953_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.40T02.2387_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.40T02.5119_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.40T02.8184_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.40T03.1623_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.40T03.5481_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.40T03.9811_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.40T04.4668_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.40T05.0119_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.40T05.6234_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.40T06.3096_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.40T07.0795_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.40T07.9433_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.40T08.9125_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.40T10.0000_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.40T11.2202_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.40T12.5893_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.40T14.1254_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.40T15.8489_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.40T17.7828_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.71T00.0631_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.71T00.0708_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.71T00.0794_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.71T00.0891_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.71T00.1000_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.71T00.1122_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.71T00.1259_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.71T00.1413_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.71T00.1585_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.71T00.1778_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.71T00.1995_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.71T00.2239_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.71T00.2512_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.71T00.2818_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.71T00.3162_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.71T00.3548_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.71T00.3981_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.71T00.4467_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.71T00.5012_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.71T00.5623_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.71T00.6310_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.71T00.7079_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.71T00.7943_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.71T00.8913_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.71T01.0000_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.71T01.1220_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.71T01.2589_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.71T01.4125_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.71T01.5849_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.71T01.7783_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.71T01.9953_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.71T02.2387_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.71T02.5119_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.71T02.8184_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.71T03.1623_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.71T03.5481_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.71T03.9811_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.71T04.4668_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.71T05.0119_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.71T05.6234_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.71T06.3096_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.71T07.0795_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.71T07.9433_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.71T08.9125_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.71T10.0000_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.71T11.2202_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.71T12.5893_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.71T14.1254_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.71T15.8489_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm0.71T17.7828_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.31T00.0631_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.31T00.0708_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.31T00.0794_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.31T00.0891_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.31T00.1000_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.31T00.1122_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.31T00.1259_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.31T00.1413_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.31T00.1585_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.31T00.1778_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.31T00.1995_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.31T00.2239_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.31T00.2512_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.31T00.2818_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.31T00.3162_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.31T00.3548_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.31T00.3981_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.31T00.4467_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.31T00.5012_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.31T00.5623_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.31T00.6310_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.31T00.7079_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.31T00.7943_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.31T00.8913_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.31T01.0000_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.31T01.1220_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.31T01.2589_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.31T01.4125_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.31T01.5849_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.31T01.7783_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.31T01.9953_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.31T02.2387_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.31T02.5119_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.31T02.8184_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.31T03.1623_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.31T03.5481_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.31T03.9811_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.31T04.4668_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.31T05.0119_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.31T05.6234_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.31T06.3096_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.31T07.0795_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.31T07.9433_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.31T08.9125_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.31T10.0000_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.31T11.2202_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.31T12.5893_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.31T14.1254_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.31T15.8489_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.31T17.7828_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.71T00.0631_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.71T00.0708_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.71T00.0794_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.71T00.0891_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.71T00.1000_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.71T00.1122_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.71T00.1259_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.71T00.1413_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.71T00.1585_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.71T00.1778_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.71T00.1995_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.71T00.2239_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.71T00.2512_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.71T00.2818_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.71T00.3162_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.71T00.3548_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.71T00.3981_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.71T00.4467_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.71T00.5012_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.71T00.5623_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.71T00.6310_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.71T00.7079_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.71T00.7943_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.71T00.8913_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.71T01.0000_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.71T01.1220_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.71T01.2589_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.71T01.4125_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.71T01.5849_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.71T01.7783_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.71T01.9953_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.71T02.2387_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.71T02.5119_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.71T02.8184_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.71T03.1623_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.71T03.5481_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.71T03.9811_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.71T04.4668_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.71T05.0119_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.71T05.6234_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.71T06.3096_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.71T07.0795_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.71T07.9433_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.71T08.9125_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.71T10.0000_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.71T11.2202_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.71T12.5893_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.71T14.1254_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.71T15.8489_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm1.71T17.7828_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm2.32T00.0631_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm2.32T00.0708_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm2.32T00.0794_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm2.32T00.0891_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm2.32T00.1000_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm2.32T00.1122_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm2.32T00.1259_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm2.32T00.1413_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm2.32T00.1585_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm2.32T00.1778_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm2.32T00.1995_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm2.32T00.2239_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm2.32T00.2512_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm2.32T00.2818_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm2.32T00.3162_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm2.32T00.3548_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm2.32T00.3981_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm2.32T00.4467_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm2.32T00.5012_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm2.32T00.5623_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm2.32T00.6310_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm2.32T00.7079_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm2.32T00.7943_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm2.32T00.8913_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm2.32T01.0000_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm2.32T01.1220_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm2.32T01.2589_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm2.32T01.4125_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm2.32T01.5849_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm2.32T01.7783_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm2.32T01.9953_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm2.32T02.2387_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm2.32T02.5119_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm2.32T02.8184_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm2.32T03.1623_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm2.32T03.5481_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm2.32T03.9811_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm2.32T04.4668_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm2.32T05.0119_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm2.32T05.6234_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm2.32T06.3096_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm2.32T07.0795_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm2.32T07.9433_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm2.32T08.9125_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm2.32T10.0000_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm2.32T11.2202_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm2.32T12.5893_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm2.32T14.1254_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm2.32T15.8489_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zm2.32T17.7828_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.00T00.0631_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.00T00.0708_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.00T00.0794_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.00T00.0891_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.00T00.1000_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.00T00.1122_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.00T00.1259_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.00T00.1413_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.00T00.1585_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.00T00.1778_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.00T00.1995_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.00T00.2239_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.00T00.2512_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.00T00.2818_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.00T00.3162_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.00T00.3548_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.00T00.3981_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.00T00.4467_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.00T00.5012_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.00T00.5623_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.00T00.6310_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.00T00.7079_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.00T00.7943_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.00T00.8913_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.00T01.0000_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.00T01.1220_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.00T01.2589_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.00T01.4125_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.00T01.5849_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.00T01.7783_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.00T01.9953_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.00T02.2387_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.00T02.5119_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.00T02.8184_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.00T03.1623_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.00T03.5481_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.00T03.9811_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.00T04.4668_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.00T05.0119_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.00T05.6234_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.00T06.3096_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.00T07.0795_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.00T07.9433_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.00T08.9125_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.00T10.0000_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.00T11.2202_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.00T12.5893_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.00T14.1254_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.00T15.8489_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.00T17.7828_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.22T00.0631_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.22T00.0708_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.22T00.0794_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.22T00.0891_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.22T00.1000_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.22T00.1122_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.22T00.1259_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.22T00.1413_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.22T00.1585_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.22T00.1778_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.22T00.1995_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.22T00.2239_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.22T00.2512_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.22T00.2818_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.22T00.3162_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.22T00.3548_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.22T00.3981_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.22T00.4467_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.22T00.5012_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.22T00.5623_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.22T00.6310_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.22T00.7079_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.22T00.7943_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.22T00.8913_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.22T01.0000_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.22T01.1220_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.22T01.2589_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.22T01.4125_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.22T01.5849_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.22T01.7783_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.22T01.9953_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.22T02.2387_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.22T02.5119_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.22T02.8184_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.22T03.1623_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.22T03.5481_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.22T03.9811_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.22T04.4668_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.22T05.0119_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.22T05.6234_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.22T06.3096_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.22T07.0795_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.22T07.9433_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.22T08.9125_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.22T10.0000_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.22T11.2202_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.22T12.5893_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.22T14.1254_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.22T15.8489_iPp0.00_baseFe.fits, gehong/data/EMILES/Ech1.30Zp0.22T17.7828_iPp0.00_baseFe.fits, gehong/data/filter/HST_WFPC2-PC.F555W.filter, gehong/data/filter/HST_WFPC2-PC.F814W.filter, gehong/data/filter/SDSS_r.filter, gehong/data/filter/SLOAN_SDSS.g.filter, gehong/data/filter/SLOAN_SDSS.r.filter, gehong/data/.DS_Store, gehong/data/AGN.NLR.fits, gehong/data/FeII.AGN.fits, gehong/data/Starlib.XSL.fits, gehong/data/fsps.nebular.fits, gehong/.DS_Store, gehong/__init__.py, gehong/config.py, gehong/cube3d.py, gehong/map2d.py, gehong/spec1d.py, src/csst_ifs_gehong.egg-info/PKG-INFO, src/csst_ifs_gehong.egg-info/SOURCES.txt, src/csst_ifs_gehong.egg-info/dependency_links.txt, src/csst_ifs_gehong.egg-info/requires.txt, src/csst_ifs_gehong.egg-info/top_level.txt, src/gehong/__init__.py, src/gehong/config.py, src/gehong/cube3d.py, src/gehong/map2d.py, src/gehong/spec1d.py, test/__pycache__/config.cpython-38.pyc, test/__pycache__/cube3d.cpython-38.pyc, test/__pycache__/map2d.cpython-38.pyc, test/__pycache__/spec1d.cpython-38.pyc, test/__pycache__/test_cube3d.cpython-38.pyc, test/__pycache__/test_spec1d.cpython-38.pyc, test/image/test_cube3d_spec.png, test/image/test_map2d_SBmap.png, test/image/test_map2d_gredmap.png, test/image/test_map2d_velmap.png, test/image/test_spec1d_agn.png, test/image/test_spec1d_ionizedgas.png, test/image/test_spec1d_singlestar.png, test/image/test_spec1d_stellarpopulation.png, test/.DS_Store, test/.coverage, test/test_cube3d.py, test/test_map2d.py, test/test_spec1d.py
src/gehong/config.py
deleted
100644 → 0
src/gehong/cube3d.py
deleted
100644 → 0
src/gehong/map2d.py
deleted
100644 → 0
src/gehong/spec1d.py
deleted
100644 → 0
test/.DS_Store
deleted
100644 → 0
File deleted
test/.coverage
deleted
100644 → 0
File deleted
File deleted
File deleted
File deleted
File deleted
File deleted
File deleted
55.4 KB
23.2 KB
26.7 KB
23.5 KB
96.9 KB
63.9 KB
170 KB